Abstract
Motivation: RNA interference (RNAi) by small interfering RNAs (siRNAs) has become a powerful tool in the fields of molecular biology and medicine. The success of RNAi gene silencing depends on the specificity of siRNAs for particular mRNA sequences. Some siRNA design guidelines have been established from siRNA sequence analysis, but designing siRNA sequences based only on these limited rules might not be effective. Experimentally validated siRNA databases have been developed over the past few years. Because of this wealth of sequence data, modules that employ machine learning methods can be developed to predict siRNA accuracy and optimize design.
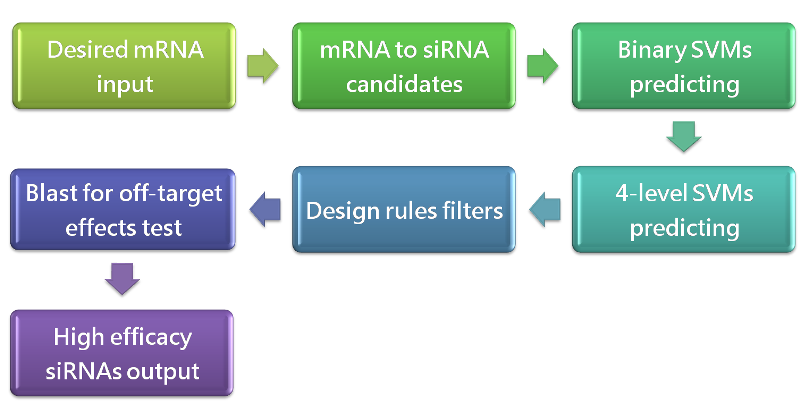
Results: In this study, we created an siRNA design tool “DEsi” that quickly selects siRNAs with high RNAi activity against a desired mRNA. DEsi combines traditional feature filters, machine learning models and BLAST to optimize siRNAs design. The prediction models in DEsi had considerable predictive power, which was validated by statistical analysis. Compared with other siRNA design tools, DEsi can quickly and accurately design siRNAs against desired mRNAs.