To manipulate protein phosphorylation prediction In the user interface of GasPhos, first the user should see a block to enter the candidate sequence as below:
The user could input the sequence in FASTA format:
After the sequence is entered, the user has to select which specific protein kinase shall phosphorylate the candidate sequence on phosphorylable amino acid residue
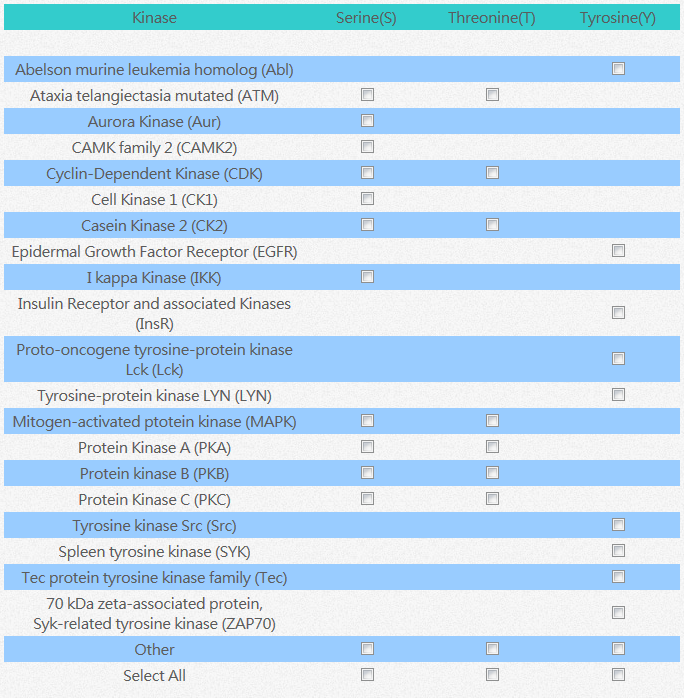
The different rows show the selections of protein kinases, and the different columns are the choice of amino acid residue the user wants to make prediction on. After entering the sequence and having those selections choose, the user could press click the "submit" button and wait for the result to show. The result of prediction shall show up in two minutes.
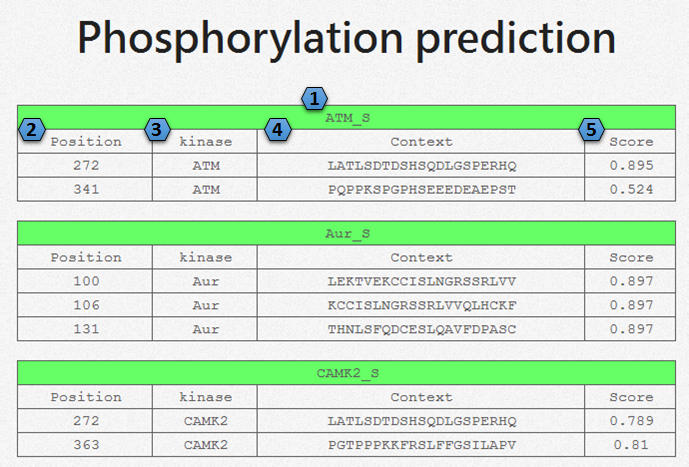
The prediction result as shown above, and the meaning of each field are as follow: