|
||||
EAT-Rice: Using machine learning approach to predicting the flanking genes expression activated by CaMV 35S enhancer in T-DNA insertion activation-tagged rice lines |
About of Research
The rice T-DNA insertion mutants from TRIM database provide important genetic resources for gene function assay. Each T-DNA insertion mutant carried four tandem repeats of CaMV 35S enhancer which could activate the expression of its flanking genes and was known as the activation-tagged mutant. Previous studies showed the expression of flanking genes were neither uniformly affected by 35S enhancer nor correlated with their distance from the 35S enhancer. Therefore, it will be a time-consuming and laborious works to check the expression of flanking genes when many genes were located near the T-DNA insertion site. In this study, we used machine learning approach to predict the expression of flanking genes in T-DNA activation-tagged mutants and used this analysis platform to further identify the function of candidate genes. The flanking genes, totally 358 genes that revealed either activated or no-activated in previous experimentally validated datasets were collected and three different regions of their DNA sequences including 1) Ups1k (one kb of upstream sequence from the start codon), 2) Distance (from the start codon of target gene to enhancer) and 3) Middle (150 bp of up- and downstream sequence around the central nucleotide of Distance region) were retrieved respectively to encode and build a two-layer machine learning prediction models. In the first layer models, the features with sequences of permutations and combinations calculating by n-gram, specific motif on promoter, nucleotides physicochemical properties and CG-island were used to build SVM models by analyzed the hidden information of three sequences. Moreover, we adopted logistical regression to estimate probability of gene activated, it was depended for weighted of feature encoding. In the second layer models, the NaïveBayes Updateable algorithm was selected from 69 classified methods to integrate this first layer models, and the system performance was 88.33% on 5 fold cross-validation, and 79.17% on independent-testing finally. To evaluate the systemic accuracy with the distance of target genes from the enhancer, results showed the accuracy was much better for genes with distance distributed between 2 to 5 kb and 10 to 20 kb respectively; and further discussed the difference of gene activated information from TRIM. Taken together, this study successfully constructed a prediction system: EAT-Rice, which could provide the expression status of flanking genes in T-DNA insertion activation-tagged mutants. |
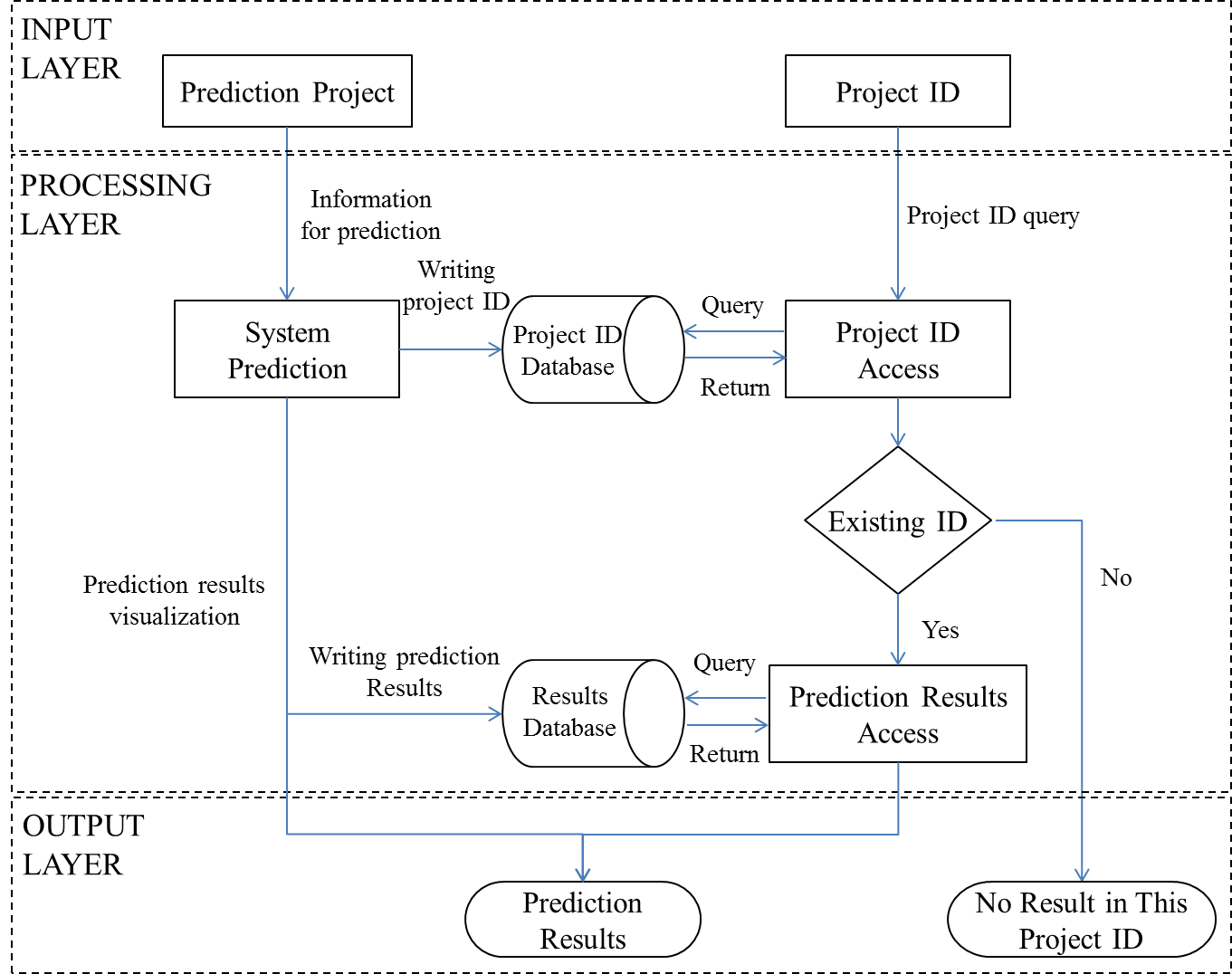
Architecture of EAT-Rice
| Natural Computing & Bioinformatics Laboratory ( NCB Lab.), Institute of Genomics and Bioinformatics, National Chung Hsing University, Taiwan. |
| For questions and comments, please contact: ywchu@nchu.edu.tw. |
| Last update for website: Jan 30, 2019 |
