WELCOME TO iStable
Mutation of a single amino acid residue can cause changes in a protein, which could then lead to a loss of protein function. Predicting the protein stability changes can provide several possible candidates for the novel protein designing. Although many prediction tools are available, the conflicting prediction results from different tools could cause confusion to users.
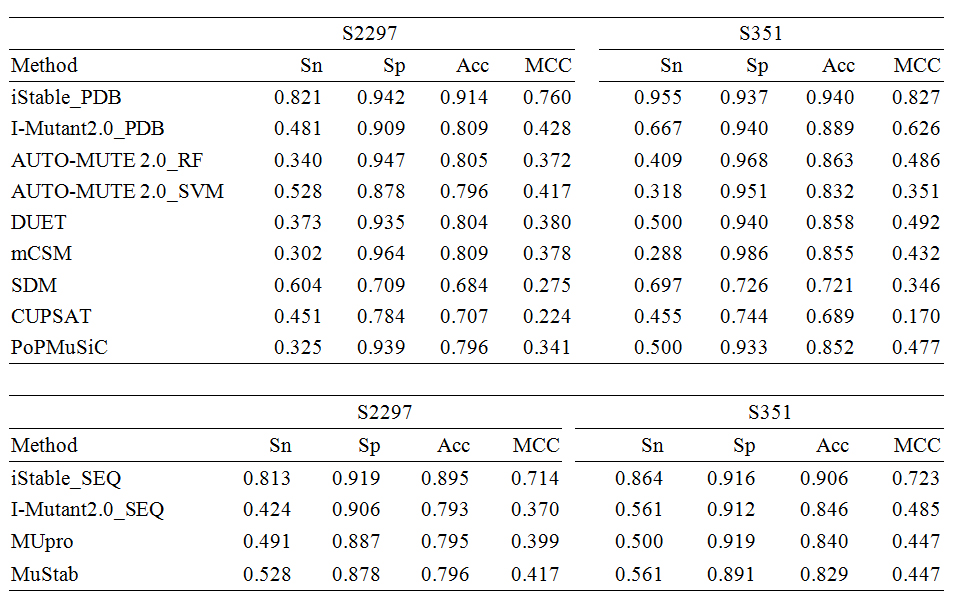
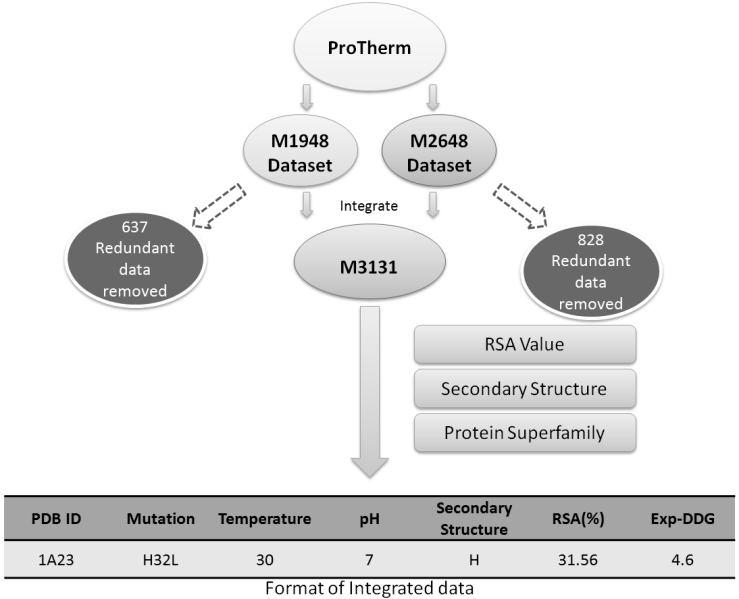
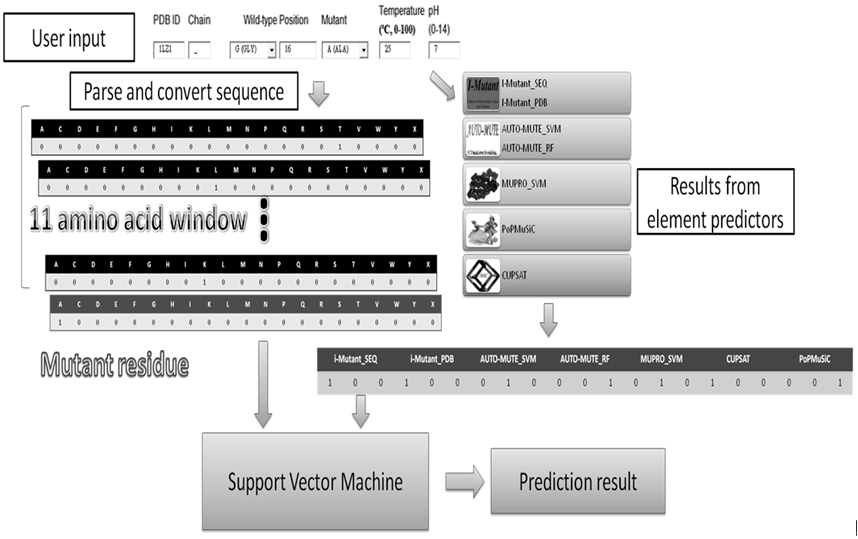
We proposed an integrated predictor, iStable, constructed by using sequence information and prediction results from different element predictors. In the learning model, iStable adopted the support vector machine as an integrator, while not just choosing the majority answer given by element predictors. Furthermore, the role of the sequence information played was analyzed in our model, and an 11-window size was determined. On the other hand, iStable is available with two different input types: structural and sequential. After training and cross-validation, iStable has better performance than all of the element predictors on several datasets. Under different classifications and conditions for validation, this study has also shown better overall performance in different types of secondary structures, relative solvent accessibility circumstances, and protein memberships in different superfamilies.
The trained and validated version of iStable provides an accurate approach for prediction of protein stability change.
Performance of iStable in S2648
 Reference:
Reference:
Chi-Wei Chen, Jerome Lin and Yen-Wei Chu* (2013) iStable: Off-the-shelf Predictor Integration for Predicting Protein Stability Changes, BMC Bioinformatics, 14(suppl 2):S5, doi:10.1186/1471-2105-14-S2-S5.
|